Bioinformatics tools aid exploration of computational biology
Science and Technology
SUMMARY: Just as amazing as what he sees, says Enke, an assistant professor of biology, are the advances in technology that make such information so accessible. To keep up with the technology advances, such as a Genome Browser developed by and hosted at the University of California, Santa Cruz, JMU's new Center for Genome and Metagenome Studies held a three-day workshop this summer on bioinformatics - the computational side of biology.
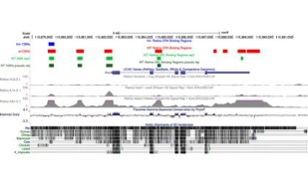
Dr. Ray Enke pulls up a chart on his computer and begins explaining what it shows:
"That’s the rhodopsin gene," he says, pointing to the colorful display. "This is the chromosome up here, that’s where the gene rhodopsin sits, that’s its neighbor gene, that’s its other neighbor gene . . . so you can visualize the data."
Just as amazing as what he sees, says Enke, an assistant professor of biology, are the advances in technology that make such information so accessible. To keep up with the technology advances, such as a Genome Browser developed by and hosted at the University of California, Santa Cruz, JMU's new Center for Genome and Metagenome Studies held a three-day workshop this summer on bioinformatics — the computational side of biology.
"We want our students to stay on the curve with all of this stuff, but in order for them to stay on the curve, we have to have faculty who know how to teach this stuff," Enke said.

While faculty made up the majority of participants — including from Eastern Mennonite High School, Radford University and Virginia Commonwealth University—students attended as well. And in addition to learning about the user friendly Genome Browser, the first two days of the workshop, supported by 4-VA at JMU, featured Data Carpentry experts and emphasized some more advanced programming.
"The Data Carpentry portion of the workshop was really focused on coding. Taking big data sets and running them through different types of analysis, but you first have to learn a set of basic coding commands," Enke said. "I have basic computing skills. Some of my colleagues, like (Dr.) Steve Cresawn (associate professor of biology), he writes his own scripts, he teaches a bioinformatics class where students are actually sitting on the command line coding in a Unix shell. All that stuff is extremely useful for studying biology, but makes people like me very anxious to even think about."
CGEMS, co-directed by Cresawn and and Dr. Jim Herrick, associate professor of biology, will soon begin planning for next summer's workshop, Enke said.
