Regulation of Gene Expression in the Vertebrate Retina
The Enke lab currently uses the chicken embryo (Gallus gallus) to study aspects of retinal developmental as well as post mortem human donor eyes to investigate retinal diseases such as age-related macular degeneration (AMD). We are interested in using two applications of Next-Generation Sequencing to better understand gene regulatory mechanisms in the developing and diseased retina. First, we are exploring the use of whole genome bisulfite sequencing (WGBS) as a means to create high-resolution maps of the epigenetic modification DNA methylation throughout the genomes of retinal cells. Second, we are using RNA-Seq to measure global mRNA gene expression in the same retinal cells. By correlating these two genome-wide data sets, we hope gain novel insights into epigenetic regulation of retina-specific gene expression.
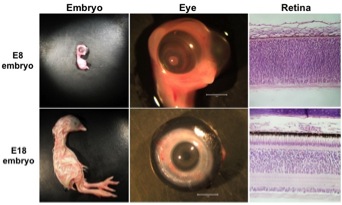
Illumina RNA-Seq analysis of retinal precursors from 8-day chicken embryos (E8) and differentiated retinal neurons from 18-day chicken embryos (E18) is being used to identify genes critical for development of the vertebrate retina.
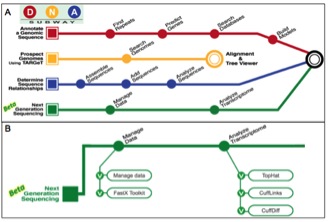
(A) The iPlant Collaborative’s DNA Subway uses the intuitive metaphor of a subway map to organize research-grade computational bioinformatics tools into simplified workflows presented in an appealing interface. (B) The Green Line of DNA Subway uses the Tuxedo Protocol, a sophisticated computational workflow incorporating open source software components for all steps required in the RNA-Seq data analysis pipeline.
